The Whole Genome Shotgun Sequencing Project of Okinawan and Australian crown of thorns starfish, Acanthaster planci.
The Whole Genome Shotgun Sequencing Project of Okinawan and Australian crown of thorns starfish, Acanthaster planci.
version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 2.0 |
 A chromosome-scale, haplotype-phased genome assembly (version: 3.2Ref, Reference Haplotype) of the coral (Takeuchi et al., 2025).
A chromosome-scale, haplotype-phased genome assembly (version: 3.2Ref, Reference Haplotype) of the coral (Takeuchi et al., 2025).
version: 3.2Ref |
 A chromosome-scale, haplotype-phased genome assembly (version: 3.2Alt, Alternative Haplotype) of the coral (Takeuchi et al., 2025).
A chromosome-scale, haplotype-phased genome assembly (version: 3.2Alt, Alternative Haplotype) of the coral (Takeuchi et al., 2025).
version: 3.2Alt |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |

version: 1.0 |
 A chromosome-scale, haplotype-phased genome assembly (version: 2.2Ref, Reference Haplotype) of the coral (Takeuchi et al., 2025).
A chromosome-scale, haplotype-phased genome assembly (version: 2.2Ref, Reference Haplotype) of the coral (Takeuchi et al., 2025).
version: 2.2Ref |
 A chromosome-scale, haplotype-phased genome assembly (version: 2.2Alt, Alternative Haplotype) of the coral (Takeuchi et al., 2025).
A chromosome-scale, haplotype-phased genome assembly (version: 2.2Alt, Alternative Haplotype) of the coral (Takeuchi et al., 2025).
version: 2.2Alt |

version: 1.0 |
version: 1.0 |

version: 1.1 |
version: |
 The whole genome shotgun sequencing project of the siphonous green alga, Caulerpa lentillifera.
The whole genome shotgun sequencing project of the siphonous green alga, Caulerpa lentillifera.
version: 1.1 |
 Genome sequencing of Chlorella sp. A99, the symbiotic algae in hydra
Genome sequencing of Chlorella sp. A99, the symbiotic algae in hydraGreen hydra harbors symbiotic Chlorella in the endodermal epithelial cells. They established mutualistic relat... version: 0.1 |
 Okinawa Mozuku Genome Project 160227
Okinawa Mozuku Genome Project 160227
version: 0.4 |

version: version 1.0 |

version: version 1.0 |

version: version 1.0 |

version: version 2 |
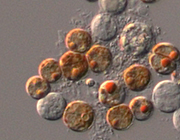 Symbiodinium clade D
Symbiodinium clade D
version: 1.0 |
 RNA sequence project of deep sea glass sponge, Euplectella aspergillum.
RNA sequence project of deep sea glass sponge, Euplectella aspergillum.
version: version 0.1 |

version: ver1.0 |
 The genome project of the lingulid brachiopod Lingula anatina.
The genome project of the lingulid brachiopod Lingula anatina. The 425-Mb genome (3,830 scaffolds with contig N50 in 56 kbp and scaffold N50 in 294 kbp) was sequenced from male gon... version: 1.0 |
 Brachiopod, Lingula anatina version 2
Brachiopod, Lingula anatina version 2
version: 2.0 |

version: 1.1 |

version: 1.1 |

The genome project of the cubozoan jellyfish Morbakka virulenta (fire jellyfish). version: |

version: version 1 |
 Nemertean, Notospermus geniculatus
Nemertean, Notospermus geniculatus
version: 2.0 |

version: catfish version 2018 |
 Phoronid, Phoronis australis
Phoronid, Phoronis australis
version: 2.0 |
 Chromosome-scale, haplotype-merged genome assembly (version: 3.0) of the pearl oyster. Genomic DNA was obtained from the MK (Mikimoto) individual (Takeuchi et al., 2022).
Chromosome-scale, haplotype-merged genome assembly (version: 3.0) of the pearl oyster. Genomic DNA was obtained from the MK (Mikimoto) individual (Takeuchi et al., 2022).
version: 3.0 |

version: 1.0 |
 The whole genome shotgun sequencing project of the acoel flatworm, Praesagittifera naikaiensis.
The whole genome shotgun sequencing project of the acoel flatworm, Praesagittifera naikaiensis.
version: 1.0 |
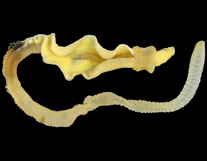 Genomic data for Hawaiian Acornworm, Ptychodera flava.
Genomic data for Hawaiian Acornworm, Ptychodera flava.
version: v3.0 |
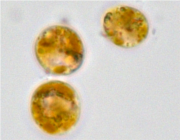 The genome project for a clade-B1 symbiodinium.
The genome project for a clade-B1 symbiodinium.
version: symb_aug_v1.120123 |
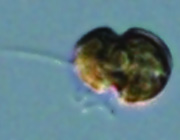 The genome project for a clade-A3 symbiodinium.
The genome project for a clade-A3 symbiodinium.
version: symbA.v1.0 |
 The genome project for a clade-C symbiodinium.
The genome project for a clade-C symbiodinium.
version: symbC.v1.0 |
 The whole genome shotgun sequence project of the Okinawan staghorn coral, Acropora digitifera.
The whole genome shotgun sequence project of the Okinawan staghorn coral, Acropora digitifera.
version: oist_v1.1 |
 Repetitive DNA (%): 34.09;
N50 (Mb): 1.39;
L50: 84;
GC content: 38.95;
Gap (%): 15.27;
Reads Coverage: 144;
Assembled size (Mb): 411;
Gene Number: 28,958;
Average gene length (bp): 1,585;
Repetitive DNA (%): 34.09;
N50 (Mb): 1.39;
L50: 84;
GC content: 38.95;
Gap (%): 15.27;
Reads Coverage: 144;
Assembled size (Mb): 411;
Gene Number: 28,958;
Average gene length (bp): 1,585;
version: |
 Repetitive DNA (%): 32.98;
N50 (Mb): 1.14;
L50: 103;
GC content: 38.93;
Gap (%): 9.75;
Reads Coverage: 158;
Assembled size (Mb): 407;
Gene Number: 30,776;
Average gene length (bp): 1,321;
Repetitive DNA (%): 32.98;
N50 (Mb): 1.14;
L50: 103;
GC content: 38.93;
Gap (%): 9.75;
Reads Coverage: 158;
Assembled size (Mb): 407;
Gene Number: 30,776;
Average gene length (bp): 1,321;
version: |
 Repetitive DNA (%): 31.92;
N50 (Mb): 1.09;
L50: 110;
GC content: 38.91;
Gap (%): 13.43;
Reads Coverage: 145;
Assembled size (Mb): 432;
Gene Number: 30,922;
Average gene length (bp): 1,306;
Repetitive DNA (%): 31.92;
N50 (Mb): 1.09;
L50: 110;
GC content: 38.91;
Gap (%): 13.43;
Reads Coverage: 145;
Assembled size (Mb): 432;
Gene Number: 30,922;
Average gene length (bp): 1,306;
version: |
 Repetitive DNA (%): 34.58;
N50 (Mb): 1.16;
L50: 103;
GC content: 38.93;
Gap (%): 7.51;
Reads Coverage: 188;
Assembled size (Mb): 408;
Gene Number: 26,445;
Average gene lengt...
Repetitive DNA (%): 34.58;
N50 (Mb): 1.16;
L50: 103;
GC content: 38.93;
Gap (%): 7.51;
Reads Coverage: 188;
Assembled size (Mb): 408;
Gene Number: 26,445;
Average gene lengt...
version: |

version: 1.0 |

version: 1.0 |

version: 1.0 |
 The whole genome shotgun sequencing project of Japanese Pearl Oyster.
The whole genome shotgun sequencing project of Japanese Pearl Oyster.
version: 1.0 + 1.1 |
 The whole genome shotgun sequencing projects of Japanese Pearl Oyster.
The whole genome shotgun sequencing projects of Japanese Pearl Oyster.
version: ver2.0 |